There are now several software packages that are capable of reducing CABB data. For the purposes of calibration, the Miriad software package is still probably the best choice. It is possible though to do a full reduction without really using Miriad, if you prefer to use the CASA package. Below is a high-level comparison of what each package is capable of doing, and this may help you decide on how to proceed.
Only Miriad can read in raw CABB RPFITS files, with the atlod task. However, CASA can use its importmiriad task to take the output of atlod and convert it into the more commonly-used MeasurementSet (MS) format.
Both packages are capable of flagging CABB data, but generally speaking, the Miriad routines (such as uvflag, blflag and pgflag) are easier to drive, and give excellent results.
Both packages can do a full calibration of the data, including flux density calibration (the latest models for 1934-638 are in CASA), and polarisation calibration. The only significant difference is that CASA gives you much more control about where different effects are considered in the calibration tables; both will give excellent calibration however.
Imaging is where the Miriad and CASA packages differ greatly. Miriad allows you to make images over wide bandwidths, and does a good job of making spectral cubes as well. It also handles making mosaicked images, but lacks the ability to do mosaicking over a wide bandwidth.
To do that properly, you will need to turn to CASA, which is also able to handle doing multiscale cleaning, and does uv-based cleaning instead of Miriad's image-based cleaning.
For a simpler but more capable imager, you may also want to consider using wsclean, which supports most of what CASA does, but is very easy to drive.
This section will describe how to get started on reducing CABB data using the Miriad software suite. More information can be found in the Miriad manual. The manual's cookbook should be read by all users before starting their first data reduction. This section will summarise how reduction differs for CABB data, and give a reasonable guide on how to do a simple continuum reduction. The Miriad manual has been updated to incorporate information on CABB data reduction.
Miriad development is mostly static, but code to fix bugs may be introduced at any time. It is therefore important that before beginning your data reduction you obtain the very latest Miriad distribution.
To get the latest Miriad distribution, please follow the instructions in the install guide. Once you have it installed on your computer, it is easy to keep up-to-date. If you installed the binary version of Miriad (which is highly recommended), you can receive updates by running
$ mirsync -u usr123
where usr123
should be replaced with your ATNF UNIX user name. If you installed from source,
then you can use the mirimport command to get updates, after which you will
need to recompile Miriad.
It is also important to report any problems you may have with your reduction
to the Miriad maintenance team so that any issues can be resolved. If
you experience any problems, please use the mirbug routine, or send an
email describing your problem to <miriad@atnf.csiro.au>. If you don't
think your problem is related to a bug, but you need advice on how best to
proceed with your reduction, please ask for help using the
ATCA Forum.
ATCA and ATNF staff routinely check
the forum and answer questions from the community, making the forum a good
place to start to look for answers as well.
To load CABB data, you will use the atlod task. This section discusses how to use this command most effectively.
For CABB data, a number of channels are affected
by self-generated interference from the 640 MHz CABB clock, and the
birdie option flags these channels during load. It also flags out
4.9% of channels on each band edge (which works out to be 100 channels for CABB's
2049 channel spectra) and, for 20/13cm data taken before 2010-Aug-17, the unusable parts of the spectrum in
the old, separate 20cm and 13cm bands. These unusable parts are due to the fact that these
receivers did not respond to all frequencies across the 2 GHz bandwidth of
CABB. This option still works for pre-CABB data as expected.
If you wish to change the number of edge channels that are flagged by the birdie
option, use the edge parameter in atlod. By default it is
set to 9.8 (per cent), which works out to 200 channels for a 2049 channel spectrum. These flagged
channels are split evenly to the top and bottom of the spectrum, so if you wanted to flag only 32
channels on each band edge for example, you should specify
edge=3.1.
The rfiflag option can be used now to automatically flag out
frequency bands that are known to be heavily affected by RFI.
This option is most useful in the 16cm band, where the ATCA
receivers are routinely affected by strong RFI. It is completely safe to use at
any band however, and should be included in the default list of options when you
use atlod. The file it uses to determine the flagging it performs can be
customised to include extra flagging (or less flagging) if so desired. By default,
this file (called rfiflag.txt) can be found in the directory
cat in the Miriad tree.
To customise it, copy it to your reduction
directory, insert or remove more frequency ranges to flag, and then load your
data with atlod.
Note that the
rfiflag option will have no effect if the birdie option
is not also specified.
Another useful atlod option is xycorr. This corrects the
measured XY phase of the ATCA, and can be safely included at all bands for
CABB data.
By default, CABB records the autocorrelations from each of the antenna that
are being used, which may not be useful if all you are interested in is the
cross-correlations required to make an image. Having the autocorrelations
present during the reduction does no harm to any of the Miriad routines,
but may become confusing and/or annoying when trying to examine the data. You
can make atlod discard these data by using the noauto option.
The other atlod options are usually band-dependent. Examples of atlod use are given below for all ATCA bands.
- 16cm band
Task: atlod in = *.C007 out = c007.uv ifsel = 1 restfreq = options = birdie,xycorr,rfiflag,noauto nfiles = nscans = nopcorr = edge =
For this band, if you used identical centre frequencies in both IFs, you need only extract one of them as both IFs will contain identical data, and keeping both may lead to confusion later on.
- 4cm band
Task: atlod in = *.C007 out = c007.uv ifsel = restfreq = options = birdie,xycorr,rfiflag,noauto,opcorr nfiles = nscans = nopcorr = 32 edge =
If you chose to use the same centre frequency for both IFs then you should extract only 1 IF using the
ifselparameter as described above. As the frequency gets higher, the need to correct for atmospheric opacity increases; we recommend correcting for opacity in 32 separate frequency bins across the band (which is the maximum number of bins to use).- 15mm & 7mm bands
Task: atlod in = *.C007 out = c007.uv ifsel = restfreq = options = birdie,xycorr,rfiflag,opcorr,noauto nfiles = nscans = nopcorr = 32 edge =
- 3mm band
Task: atlod in = *.C007 out = c007.uv ifsel = restfreq = options = birdie,xycorr,rfiflag,noauto nfiles = nscans = nopcorr = edge =
In the 3mm band, sky temperature (and thus opacity) is determined by the use of the paddle scan, so there is no need to include opacity corrections in the loading process.
For higher frequency data, the atfix routine may be required to apply corrections to the antenna positions and system temperature measurements.
During a baseline reconfiguration at the ATCA, staff perform a baseline calibration (to determine precisely where each antenna is with respect to each other) at 5 GHz and 17 GHz. During normal observing, the solution determined at 5 GHz is applied on-line, and is usually very good. However, for mm data, you may wish to use more precise antenna positions obtained from the 17 GHz baseline calibration. This may aid in phase referencing experiments.
The atfix routine can “fix” the positions of the ATCA antennas based on the solution file that is made available by the ATCA staff on the baseline solutions page. The high-frequency baseline solutions are not routinely computed for each reconfiguration, so if you'd like to have one, please contact ATCA staff with your request.
The task atfix is also highly recommended for the reduction of 3mm data.
During an observation at 3mm, the system temperature is determined using the
paddle during a so-called “paddle scan”. These scans are usually tens of
minutes apart, during which time the system temperature recorded by the
correlator does not change. To make Miriad interpolate the system
temperature between the paddle scans, use atfix with default parameters,
or with tsyscal=interpolate explictly set.
Task: atfix vis = 3mm_data.uv select = out = 3mm_data_fixed.uv dantpos = tsyscal = options = array =
After loading the data into a Miriad dataset, it is usually most convenient to split this into many datasets, each containing an individual source at a single central frequency. This is done using the uvsplit task.
At its simplest, it can be run with only the vis parameter set to
the parent dataset, but it is also very easy to use the select parameter
here to discard data that was affected by antenna shadowing:
Task: uvsplit vis = c007.uv select = -shadow(22.5) options = maxwidth =
The select keyword can also be used to split out only certain sources,
or to effectively flag out certain antenna. Read about the select
keyword
in the Miriad manual.
There are two options that are useful here. Specify the
mosaic option if you are splitting a dataset that contains a mosaic
observation, and specify the clobber option if you want to overwrite
any previously split-out datasets in the current directory.
The maxwidth parameter is new to the CABB era, and allows you to
break up the 2 GHz CABB bandwidth into smaller chunks. This will be most useful
at lower frequencies where 2 GHz represents a large fractional bandwidth. At
such frequencies, calibration and imaging is far more challenging than at higher
frequencies. The bandwidth specified here should be specified in GHz. For example,
to mimic the maximum bandwidth available from the old correlator, you could
specify maxwidth=0.128 here. Recent experience shows
however that excellent calibration can be obtained using the entire 2 GHz band simultaneously, even
in the 16cm band. Splitting up the band at this point then is only recommended for those who have
an actual need to do so.
In most cases, if the birdie and rfiflag options are given
to atlod, the data will be mostly free from the most destructive RFI at this point. However,
you should always examine your data with the uvplt and uvspec tasks
to make sure.
Using uvplt, examining amplitude and phase versus time is a good way to
find time ranges that have been affected by RFI. Ensure that you specify
options=nofqav to prevent uvplt from averaging your data over
frequency before plotting. Any RFI present in your observations may
be restricted to only a few channels, and these channels can be identified
by using uvspec.
To flag this data, you can use the following tasks.
- uvflag
Use the
selectandlinekeywords to select the bad data and setflagval=flagto flag the data as bad. uvflag can be used to flag any set of bad data, but can be tediously repetitive.- blflag
This task can replicate many of the plots that uvplt generates, which may allow you to directly select the outliers representing bad data.
- pgflag
This task replaces the old tvflag task that no longer works on displays with more than an 8-bit colour depth. pgflag displays data as a channel-time waterfall plot and allows direct selection of ranges of bad data and flexible flagging options. It also includes an automatic flagging routine that uses the SumThreshold method that is available in the AOFlagger, and is highly recommended for use during continuum reductions.
One trap that you may find is that if you are observing at 3mm and some other band in the same run, and you elected to keep CA06 tracking for the 3mm observations, the data from CA06 will not automatically be flagged. This is because CA06 has almost all the systems required for 3mm observing (save for the electronics), and so the antenna really is “on source” according to the telescope. You should use uvflag to flag out CA06 data in this case.
Automatic flagging with pgflag can be very effective if it is done in the right way. It is most useful for continuum observations in any band, and for spectral line observations when the line is not detectable in a single integration. If your spectral line is bright enough to be detected in a single integration, then the flagging routine will likely identify it as RFI.
The pgflag task accepts a number of parameters that control the automatic flagging process. Craig Anderson identified, through trial and error, a recipe that works quite well for 16cm data; this recipe is described in an ATCA Forum post Craig started. We describe this recipe here as well, and we recommend this recipe for data in any band.
This flagging recipe uses the concept that the polarisation properties of interference are quite different to that of the signals we're interested in. For the most part, the radio sky is free of discrete sources of circular polarisation, and thus most measurements of Stokes V from our interferometer should be thermal noise only. This assumption holds over all frequencies as well. So if we see a large deviation from thermal noise in Stokes V, and this deviation is confined to a narrow range of frequencies, the most likely culprit is RFI. The first step of the flagging recipe then is to look for such signals in Stokes V and flag all the visibilities over the frequencies and times they are seen. To do this with pgflag:
$ pgflag stokes=i,q,u,v flagpar=8,5,5,3,6,3 "command=<b"
This flagging can be done on each individual source after it has been split out. Of course, because
the output of the interferometer should not vary for Stokes V regardless of the field being observed, you can
also flag a single IF all together. That is, you can take a dataset output by uvsplit
with options=nosource and flag it directly without further splitting it into
individual sources. You can also do the flagging before and after calibration. Indeed, you may
have success in doing an initial round of Stokes V flagging before trying to mfcal
the bandpass calibrator, and another round of flagging with the same command after
calibration.
As an illustration, Figure 4.1 and Figure 4.2 show what pgflag will display when told to show Stokes V from a dataset centred at 2100 MHz (a typical 16cm dataset) with fourteen scans on nine different sources, obtained in October 2015. Channel number is shown along the horizontal access, and as the 16cm band is presented to the correlator as a lower sideband, higher channel numbers correspond to lower frequencies. The time of the observation increases along the vertical axis towards the top of the image and the amplitude of each channel is represented as a grey scale which is outlined in purple toward the right of the image. Flagged regions are shown as completely black. Figure 4.1 shows a 60m baseline and Figure 4.2 shows a 4469m baseline. The only flagging done to this dataset is at the edges to remove the roll-offs, which removes 10.5% of the data on each baseline, and the data are uncalibrated. These two figures clearly show how much more sensitive short baselines are to RFI than long baselines.
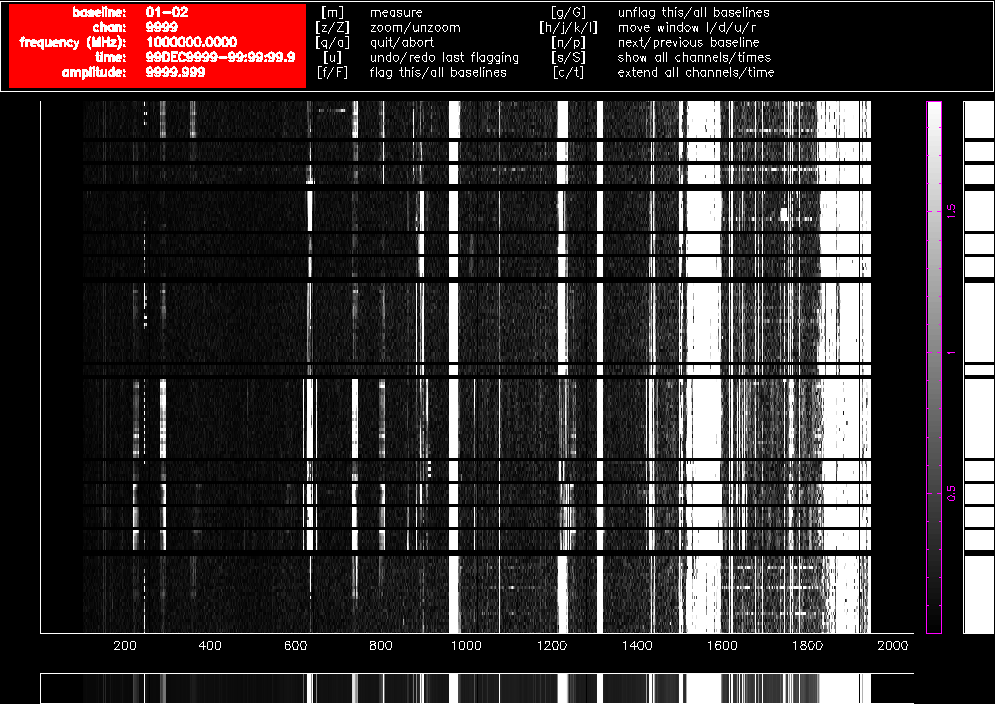
Figure 4.1. A pgflag display showing Stokes V data on a short baseline in a 16cm dataset before flagging.
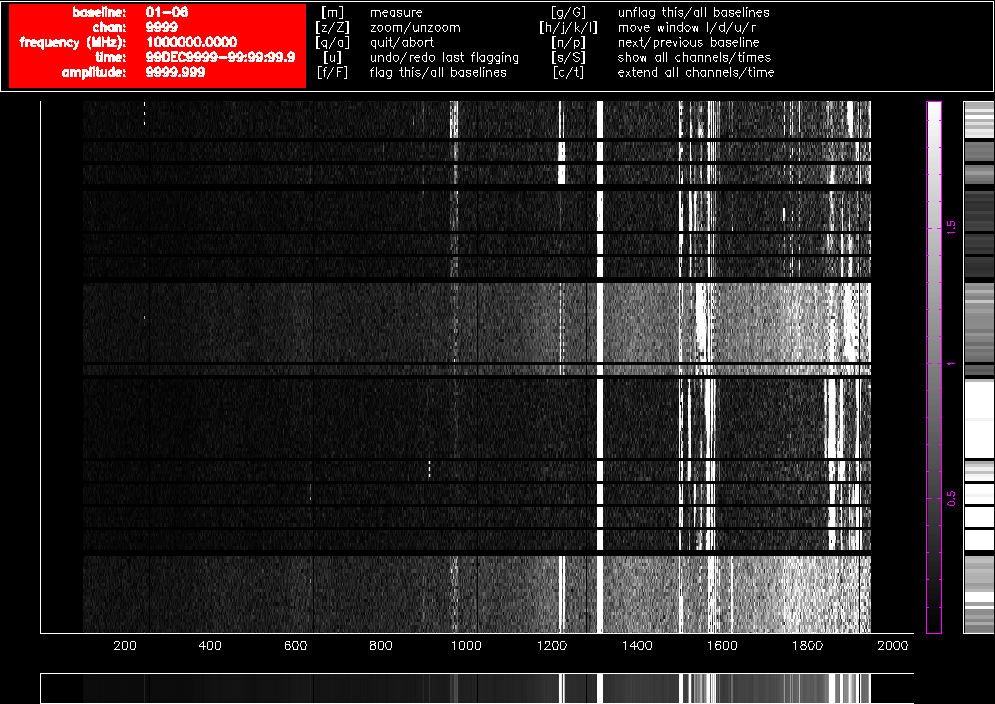
Figure 4.2. A pgflag display showing Stokes V data on a long baseline in a 16cm dataset before flagging.
After using pgflag as described above on the uncalibrated data, then bandpass calibrating and flagging with pgflag in the same way again, the data on the same baselines look like Figure 4.3 and Figure 4.4. Approximately 40% of the short baseline data is now flagged, and approximately 25% of the long baseline data.
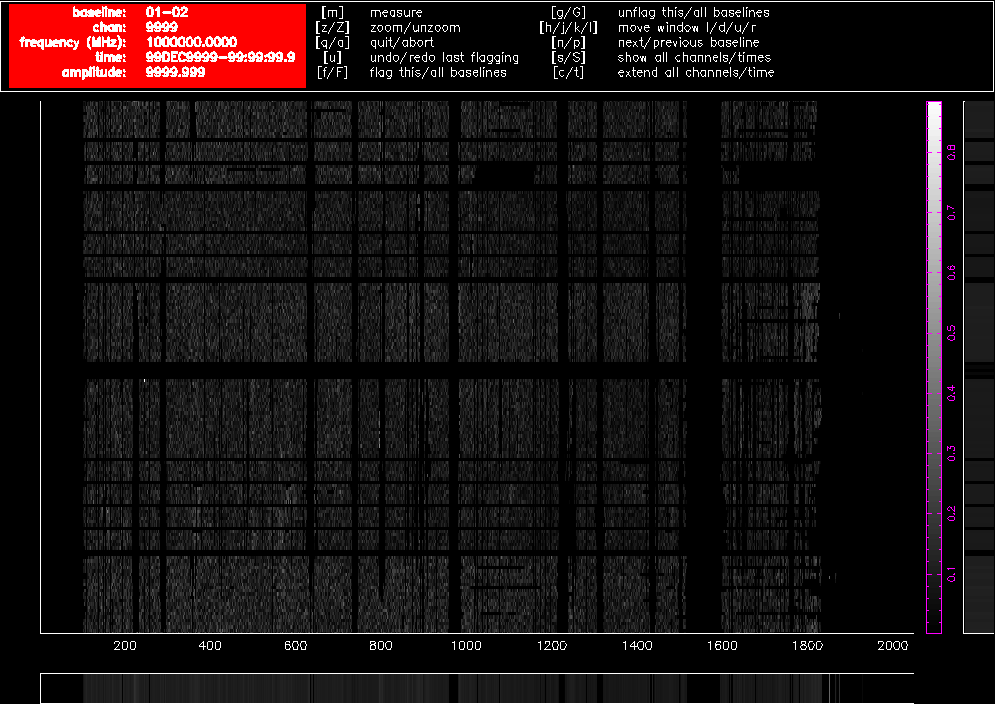
Figure 4.3. A pgflag display showing Stokes V data on a short baseline in a 16cm dataset after flagging and bandpass calibration.
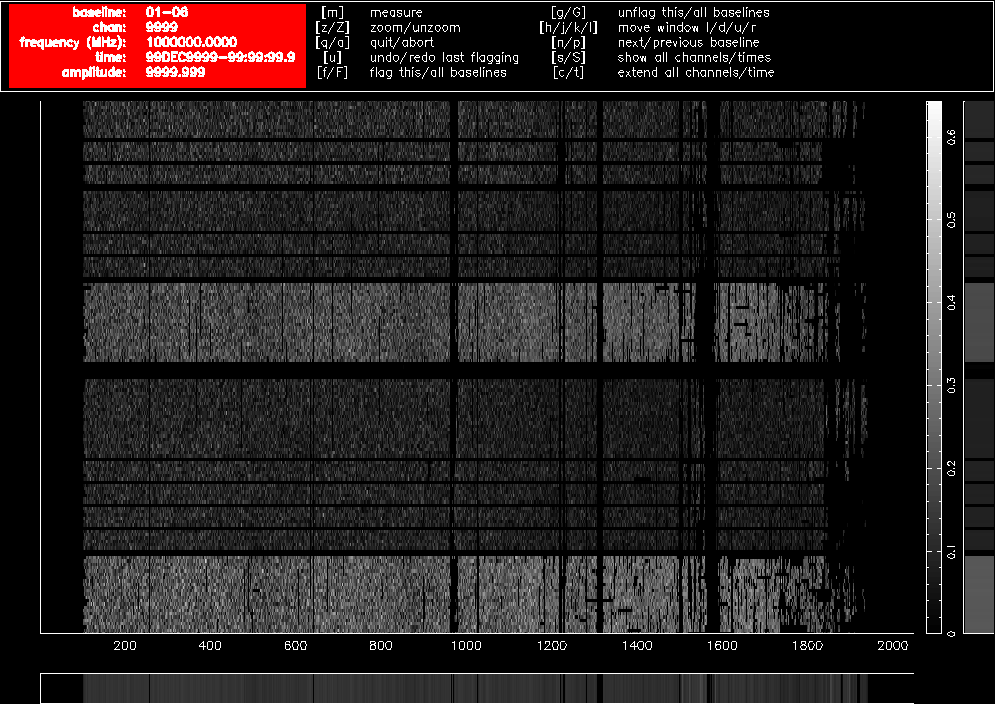
Figure 4.4. A pgflag display showing Stokes V data on a long baseline in a 16cm dataset after flagging and bandpass calibration.
As you can see, no obviously wrong signals can be identified in Stokes V after this process, but that does not necessarily mean that all RFI has been deleted from the data. Figure 4.5 and Figure 4.6 show Stokes Q on the same baselines as before, after the Stokes V flagging and bandpass calibration, and further RFI is obvious in those figures. We now flag using the linear-polarisation Stokes parameters Q and U.
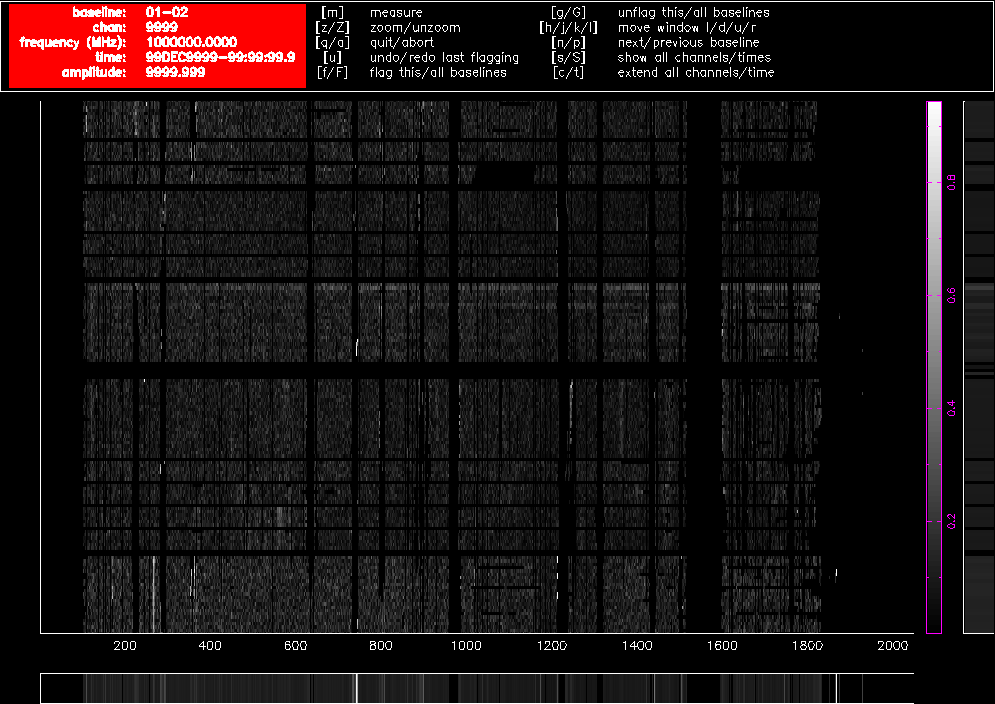
Figure 4.5. A pgflag display showing Stokes Q data on a short baseline in a 16cm dataset after flagging in Stokes V and bandpass calibration.
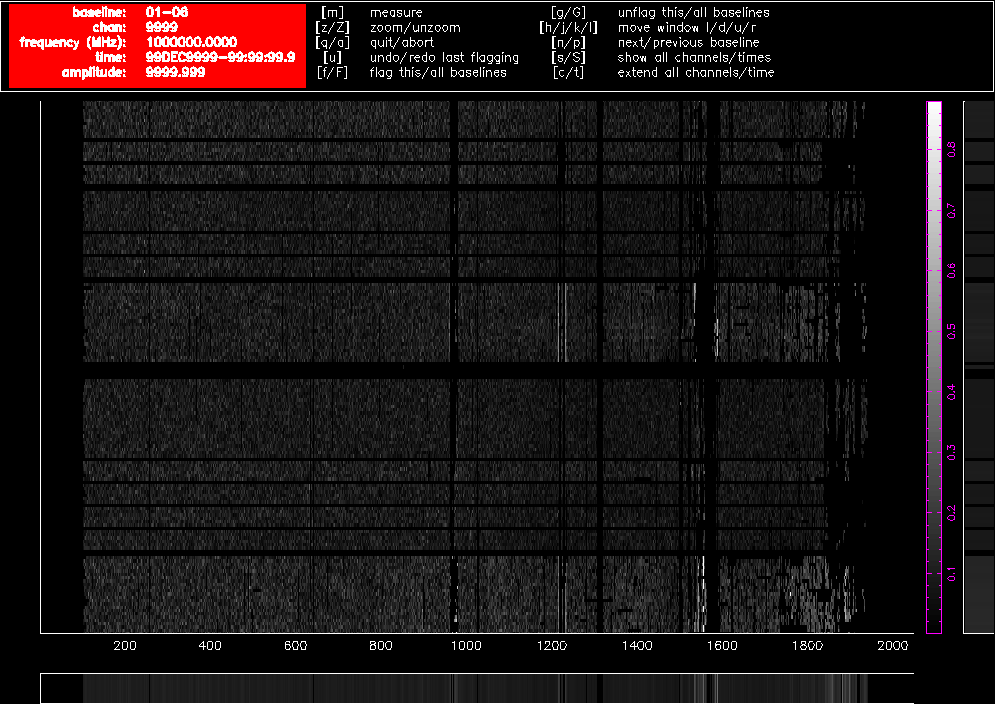
Figure 4.6. A pgflag display showing Stokes Q data on a long baseline in a 16cm dataset after flagging in Stokes V and bandpass calibration.
To do this we use pgflag as follows. First, flag Stokes Q:
$ pgflag stokes=i,v,u,q flagpar=8,2,2,3,6,3 "command=<b"
And then Stokes U:
$ pgflag stokes=i,v,q,u flagpar=8,2,2,3,6,3 "command=<b"
Again, these flagging commands can be done on each source individually, or on the dataset as a whole.
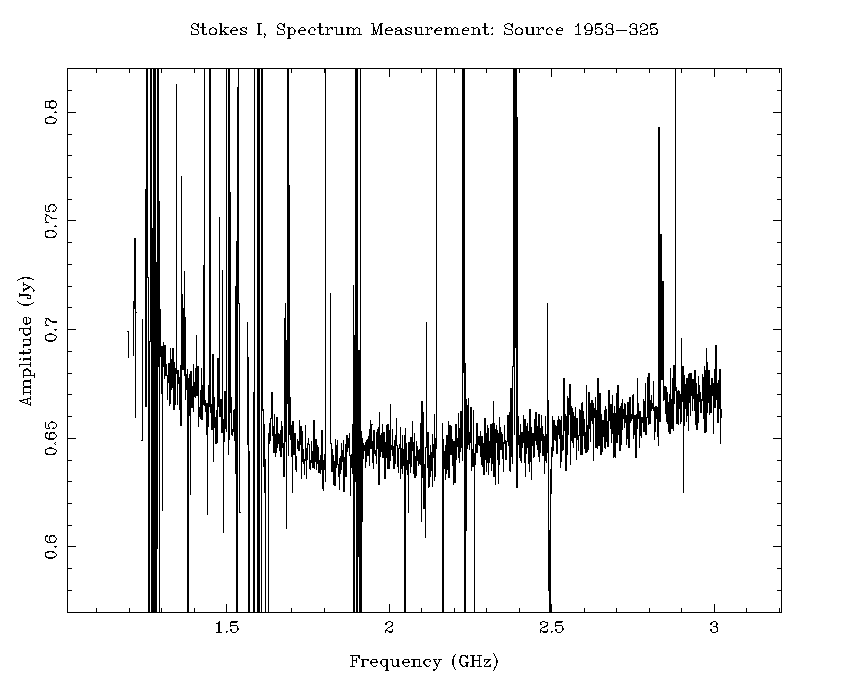
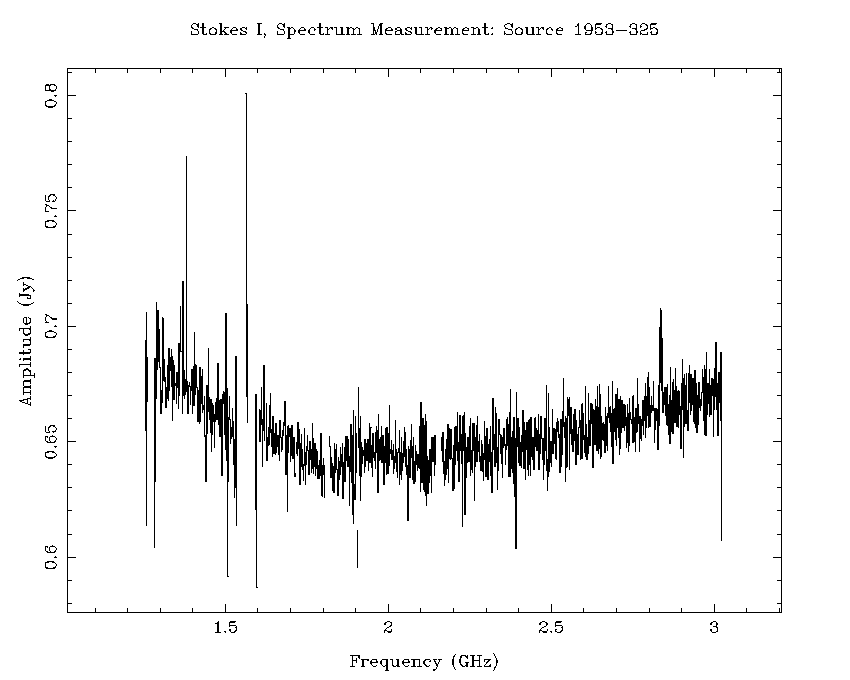
An example of how effective this flagging recipe can be is shown in Figure 4.7 and Figure 4.8. Using the calibration solutions derived from the flagged dataset, we can compare the 16cm spectrum of the calibrator 1953-325 before any flagging (Figure 4.7) to the spectrum after flagging (Figure 4.8). After the flagging process, only a small amount of RFI remains, and this is not likely to cause problems when measuring the continuum spectrum. On the shortest baselines, between 45-50% of the data has been flagged, but on the baselines to antenna 6 less than 30% of the data needed to be flagged. In terms of channels, 1625 of the channels have usable data out of the 2049 total channels, representing a spectral coverage loss of approximately 21%.
The calibration strategy for ATCA data has been refined for the CABB era, to ensure accurate calibration of wide bandwidths. This strategy also has the benefit of being applicable to the vast majority of ATCA data with little alteration. It is described best in the Miriad cookbook, but it will also be summarised here (although not explained in the same depth).
Calibration begins by determining the bandpass correction, using mfcal with
your bandpass calibrator. To prevent rapid gain variations from being entangled with your bandpass
solution, the interval parameter should be set to the cycle time (which is
usually 0.1 minutes). Since your bandpass calibrator should be very bright, such a short interval
should still have plenty of signal-to-noise. By default, mfcal uses
CA03 as the reference antenna, but if that antenna wasn't available during the observations,
ensure that you set refant to be an antenna that was available.
If you are reducing 3mm data, and CA02 was available, you should set it as the reference antenna. You should do this because it has the only noise diode in the array at 3mm, and setting it as the reference antenna may make it easier to do polarisation calibration.
| cm reduction | mm reduction |
|---|---|
Task: mfcal vis = 1934-638.2100/ line = stokes = edge = select = flux = refant = minants = interval = 0.1 options = tol = |
Task: mfcal vis = 1921-293.17000/ line = stokes = edge = select = flux = refant = minants = interval = 0.1 options = tol = |
Bandpass calibration in the cm bands (16 & 4) is usually done with the ATCA primary flux density calibrator 1934-638, as it has a large flux density at these frequencies. Since the flux density of 1934-638 is known as a function of frequency, the bandpass solution obtained in these bands is very good and takes into account how 1934-638 varies across the band. But if you use another calibrator for which Miriad doesn't have a flux density model, then the bandpass solution will be dependent on how that calibrator's flux density varies with frequency, which is a bad situation. You will need to correct for this, and how to do this correction will be described later on.
After bandpass calibration, the next step is to do some time-variable gain and polarisation calibration. If the bandpass calibrator is 1934-638, or if the bandpass calibrator has been observed several times during the run over a large range of parallactic angle, then you can use it for this step. Otherwise you should use the gain calibrator you observed.
| cm reduction | mm reduction |
|---|---|
Task: gpcal vis = 1934-638.2100/ select = line = flux = refant = minants = interval = 0.1 nfbin = 2 tol = xyphase = options = xyvary |
Task: gpcopy vis = 1921-293.17000 out = 2243-123.17000 mode = options = Task: gpcal vis = 2243-123.17000/ select = line = flux = refant = minants = interval = 0.1 nfbin = 2 tol = xyphase = options = xyvary,qusolve |
You should use an nfbin of at least 2 for all bands. This will
allow gpboot to correct any linear slope errors present in the
bandpass table. This also makes it unnecessary to use mfboot
unless your flux density calibrator is a planet. You should also increase the
interval parameter if the calibrator is not bright enough to
give enough signal-to-noise with 0.1 minutes of integration time.
If you used 1934-638 in the mfcal and gpcal steps, then you already have a flux calibrated dataset that can be used to fix the flux density scale on the other datasets.
You should now copy the solutions from the 1934-638 dataset to the gain calibrator dataset using gpcopy, and determine the time-variable gains with gpcal.
Task: gpcopy vis = 1934-638.2100/ out = 2243-123.2100/ mode = options = Task: gpcal vis = 2243-123.2100/ select = line = flux = refant = minants = interval = 0.1 nfbin = 2 tol = xyphase = options = xyvary,qusolve
The gpcal step is likely to make the flux density scale on your gain calibrator slightly different to the true scale. To fix this you should use gpboot.
Task: gpboot vis = 2243-123.2100/ cal = 1934-638.2100/ select =
You can now be confident that your gain calibrator is properly flux calibrated, and can now proceed to use it to analyse your science target.
At this point in the mm reduction, you need to do the flux density calibration. How this happens depends on whether you're using a planet as the flux density calibrator, or 1934-638. After copying the calibration tables from the calibrator used in the gpcal step to the flux density calibrator, you use gpcal followed by gpboot if you're using 1934-638, or mfboot for planets.
| 1934-638 flux calibrator | planetary calibrator |
|---|---|
Task: gpcopy vis = 2243-123.17000/ out = 1934-638.17000/ mode = options = Task: gpcal vis = 1934-638.17000/ select = line = flux = refant = minants = interval = 0.1 nfbin = 2 tol = xyphase = options = xyvary,qusolve,nopol Task: gpboot vis = 2243-123.17000/ cal = 1934-638.17000/ select = |
Task: gpcopy vis = 2243-123.33000/ out = uranus.33000/ mode = options = Task: mfboot vis = 2243-123.33000,uranus.33000/ line = select = source(uranus) flux = mode = clip = device = /xs options = |
Note the use of the nopol option for the options
parameter in gpcal, when 1934-638 is being used as the flux density calibrator.
This option tells gpcal that it should accept the leakage solution
obtained from the gain calibrator, which should be good since the gain calibrator is likely
to have much greater hour-angle coverage than the flux density calibrator.
In May 2016, Miriad's high-frequency flux density scale for 1934-638 was
changed to align the ATCA's scale to that of the VLA and the
absolute scale used by Planck. See Section 2.2.4
for details about this new scale. By default, after May 2016, Miriad
uses this new scale when setting the flux density of 1934-638 for frequencies above 11 GHz. The flux
densities measured from reductions using pre-May 2016 and post-May 2016
Miriad will therefore differ for observations above 11 GHz. If you would
like to keep using the pre-May 2016 flux density scale for 1934-638, you should use the
oldflux option that is available in the mfcal,
and gpcal commands.
At this point the gain calibrator will be reasonably well calibrated. If you are not especially concerned about flux density calibration accuracy, you can proceed to analyse your science target.
If you want to flux calibrate your data as well as possible, then you can use data from the other simultaneously observed band to improve your solution. Basically, the bandpass solution you made earlier assumed that the bandpass calibrator had a constant flux density with frequency (i.e. a flat-spectrum source). Because of this, the bandpass solution will likely be in error. Our gain solutions with two bins will have corrected this somewhat, but we can also try to emulate the situation in the cm band, and provide mfcal with a model of the flux density of the bandpass calibrator.
To do this, we copy the gain solutions from the gain calibrator back to the bandpass calibrator, recalibrate, and bootstrap the flux density scale.
Task: gpcopy vis = 2243-123.17000/ out = 1921-293.17000/ mode = options = Task: gpcal vis = 1921-293.17000/ select = line = flux = refant = minants = interval = 0.1 nfbin = 2 tol = xyphase = options = xyvary,qusolve,nopol Task: gpboot vis = 1921-293.17000/ cal = 2243-123.17000/ select =
Note that these steps don't change regardless of the type of flux density calibrator you are using.
You should do the same calibration process with the other simultaneously observed band. At the end of the process you should have two bandpass calibrator datasets, each being flux density calibrated via the gain calibrator. You can now measure the flux density of the bandpass calibrator over the entire range of observed frequencies, using the uvfmeas task. The principle here is that even if the slope over a single band is wrong due to the incorrect bandpass calibration assumption, fitting a flux density model using two independently-calibrated bands should lower the uncertainties and provide a very good model of the actual flux density of the source.
Task: uvfmeas vis = 1921-293.17000/,1921-293.19000/ select = line = stokes = i hann = offset = order = 1 options = plotvec,log,mfflux yrange = device = /xs nxy = log = fitp = feval =
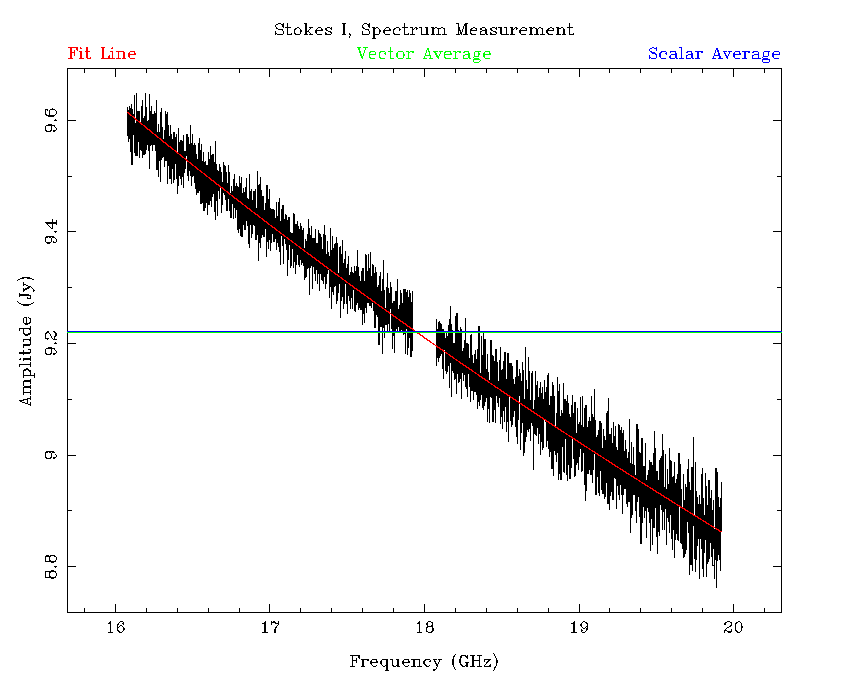
The uvfmeas task does a linear least-squares fit to the log of the vector-averaged Stokes I flux density as a function of the log of the frequency. The plot it makes will show the averaged data in both bands, and the line of best fit (Figure 4.9). It will also output a string like:
MFCAL flux= 9.4126, 17.0,-0.3801
The string is usable as the flux in mfcal,
which will fit the bandpass solution while assuming the flux density model we just
determined. The image in Figure 4.9 is actually
the result of this two stage process, and you can see the excellent alignment
between the two bands, which are independently processed.
Once you have measured the flux density model for the bandpass calibrator, you must redo the calibration, but the bandpass calibrator becomes the flux density calibrator. The process is slightly different as well, and is summarised below.
Task: mfcal vis = 1921-293.17000/ line = stokes = edge = select = flux = 9.4126,17.0,-0.3801 refant = minants = interval = 0.1 options = tol = Task: gpcopy vis = 1921-293.17000/ out = 2243-123.17000/ mode = options = Task: gpcal vis = 2243-123.17000/ select = line = flux = refant = minants = interval = 0.1 nfbin = 2 tol = xyphase = options = xyvary,qusolve Task: gpcopy vis = 2243-123.17000/ out = 1921-293.17000/ mode = options = Task: gpcal vis = 1921-293.17000/ select = line = flux = refant = minants = interval = 0.1 nfbin = 2 tol = xyphase = options = xyvary,qusolve,nopol Task: gpboot vis = 2243-123.17000/ cal = 1921-293.17000/ select = Task: mfboot vis = 2243-123.17000,1921-293.17000/ line = select = source(1921-293) flux = 9.4126,17.0,-0.3801 mode = clip = device = /xs options =
At this point both the bandpass and gain calibrator datasets will be very accurately flux density calibrated. Simulations suggest that this two-stage process can improve the flux density calibration accuracy to the 1% level for the bandpass calibrator, and to around the 3% level for any other calibrators referenced to it. Observations have confirmed this in the 15mm band, while for the 7mm band the flux calibration accuracy for an arbitrary gain calibrator is around 10%.
The calibration technique outlined above is robust and generally applicable, but may not work adequately for your data without alteration, and will almost certainly not work in an automated fashion. You should always be prepared to iterate between flagging and calibration, and you should use the visualisation tasks uvplt, uvspec and closure to check that the calibrator visibilities look as they should. You can also look at the calibration solutions directly using the gpplt task. Using uvfmeas to view the calibrators in both simultaneously-observed bands may also give you confidence that your calibration is good, or otherwise.
Imaging of CABB data can be performed in exactly the same way as pre-CABB data, however you may get better results with a slightly different method.
For high-frequency observations, where a bandwidth of 2 GHz does not represent a very large fraction of the sky frequency, then the imaging strategy used for pre-CABB data will most likely be adequate. For lower frequencies the multi-frequency strategies are more likely to produce good results. In particular, the use of the mfclean task should be considered. Read the excellent description of why mfclean is necessary in the Miriad cookbook to see if you will need to proceed with this strategy.
If you don't need mfclean then you should proceed with the usual invert, clean and restor method of creating an image, and combine this with selfcal or gpscal if appropriate. In this case, nothing should be different from pre-CABB reduction.
If you do need mfclean then the inputs to invert will need to be
slightly different as well. Firstly, both the mfs and sdb
options will need to be passed to invert. mfclean also requires
that the beam be three times the size of the region being cleaned, which can be quickly specified
in the latest version of Miriad in the imsize.
Task: invert vis = obs.uv map = obs.map beam = obs.beam imsize = 3,3,beam cell = offset = fwhm = sup = robust = 0.5 line = ref = select = stokes = i options = mfs,sdb mode = slop =
The first two options to the imsize parameter are usually the
number of pixels in the x and y direction respectively that the image will have. But when
you specify the beam option as the third argument to
imsize, the first two options are interpreted as multiples of the
FWHM of the primary beam at the central frequency of the image. In the example above, both the
dirty image and the synthesised beam image would cover 3 times the primary beam FWHM in each
direction. In earlier versions
of Miriad, both the imsize and
cell parameters would have needed to be determined manually and
explicitly set to do the same thing.
With the synthesised beam covering 3 times the primary beam FWHM, mfclean can be used to clean the entire primary beam.
Task: mfclean map = obs.map beam = obs.beam model = out = obs.clean gain = cutoff = niters = region = percentage(33) minpatch = speed = mode = log =
Here we are selecting the central third of the image (the primary beam
area) with the region keyword. This also satisfies the
mfclean requirement that a guard band be present around the
region being cleaned. The other parameters are very similar to those
of clean and should be set as usual to obtain good results.
After mfclean has run, restor should be used in the normal way to produce the cleaned image. selfcal can also be used in the usual fashion in this process.